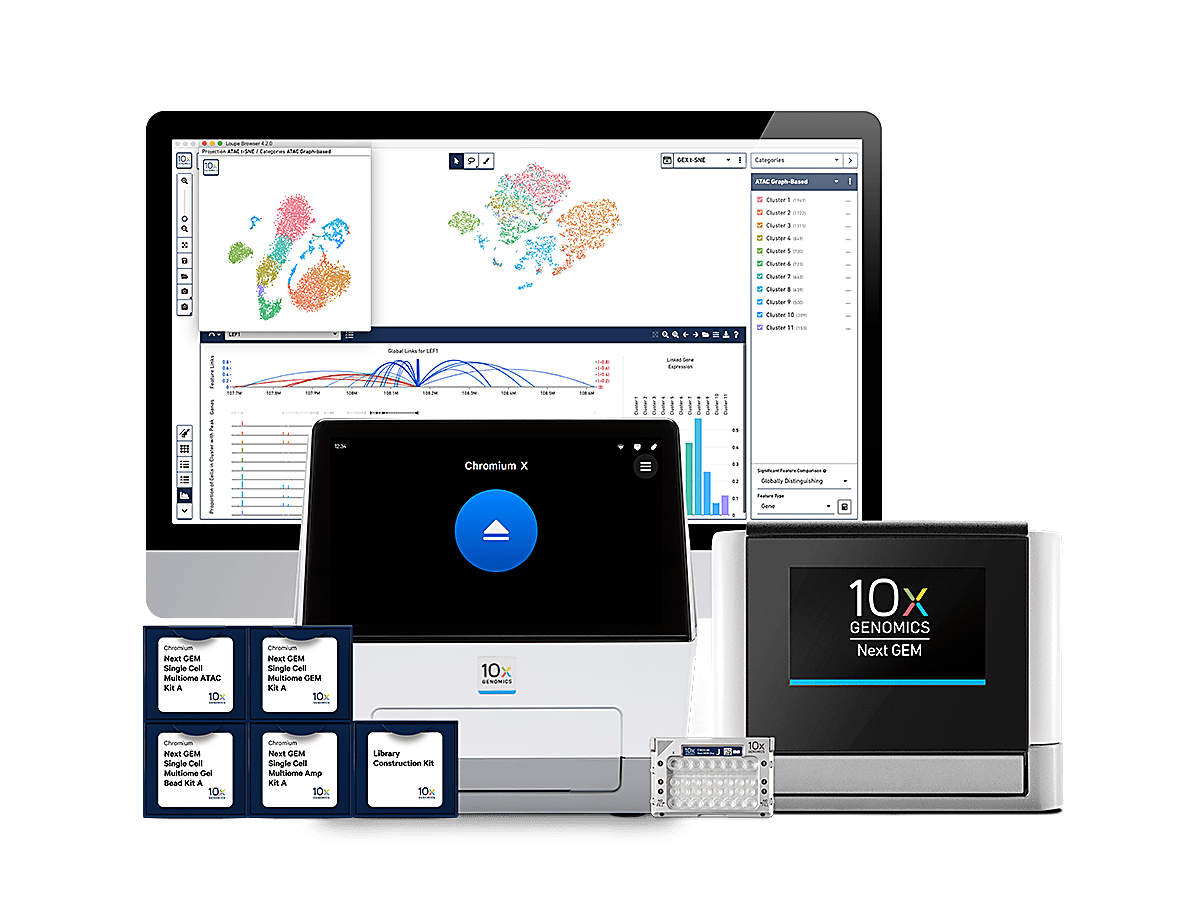
One cell, two readouts
Simultaneous detection of mRNA and chromatin accessibility from the same cell.
Define gene regulation
Discover new gene regulatory interactions.
Single cell resolution
Detect rare cell populations with single cell precision.
Flexible and scalable
Profile hundreds to tens of thousands of cells per sample.
Streamlined data analysis
Explore gene expression and chromatin accessibility simultaneously with easy-to-use software.
Efficient lab workflow
From sample to sequencing-ready library in two days.
What you can do
Deeper characterization of cell types and states
Characterize complex cell populations and capture cellular heterogeneity to uncover hidden insights with combined epigenomic and gene expression profiling. Leverage gene expression markers to more easily interpret epigenetic profiles.
Discover new gene regulatory interactions
Combine discovery of regulatory elements with gene expression to explore gene regulatory interactions driving cell differentiation, development, and disease.
Make the most of your precious samples
Maximize insights from your limited samples by obtaining transcriptome and open chromatin information from the same cell.
Explore disease complexity
Understand how gene regulatory networks are disrupted in disease. Interrogate epigenetic remodeling and mechanisms of therapeutic resistance.
Proven Results
Resources
Find out the latest resources for Chromium Single Cell Multiome ATAC + Gene Expression.
Identification of a tumor–specific gene regulatory network in human B-cell lymphoma
Identification of a tumor–specific gene regulatory network in human B-cell lymphoma
Data Spotlight, 10x Genomics
Data Spotlight, 10x Genomics
Grant Application Resources for Single Cell Multiome ATAC + Gene Expression
Grant Application Resources for Single Cell Multiome ATAC + Gene Expression
Grant Application Resources, 10x Genomics
Grant Application Resources, 10x Genomics
Massively parallel simultaneous profiling of the transcriptomic and epigenomic landscape at single cell resolution
Massively parallel simultaneous profiling of the transcriptomic and epigenomic landscape at single cell resolution
Poster, 10x Genomics
Poster, 10x Genomics
Our End-to-End Solution

Chromium Instrument with Next GEM technology
Our scalable instrument
Our compact instrument

Chromium Single Cell Multiome ATAC + Gene Expression Reagents
With our reagent kits, simultaneously detect gene expression and open chromatin regions from the same single cell and generate ready-to-sequence NGS libraries.
Analysis and Visualization Software
Cell Ranger ARC, our analysis pipeline and Loupe Browser, our visualization software.

World-Class Technical and Customer Support
Our expert support team can be contacted by phone or email at support@10xgenomics.com
Workflow
- 1
Prepare your sample
Start with a nuclei suspension isolated from cell culture, primary cells, or fresh or frozen tissue.
CHROMIUM NUCLEI ISOLATION KIT
For a wide range of frozen mammalian tissues, the Chromium Nuclei Isolation Kit offers an all-in-one kit for easy and reliable nuclei isolation optimized for the 10x Genomics Single Cell Multiome assay.
Resources - 2
Construct your 10x library
Construct a 10x barcoded library using our reagent kits and a compatible Chromium instrument. Each member of the Chromium instrument family encapsulates each cell with a 10x barcoded Gel Bead in a single partition. Within each nanoliter-scale partition, cells undergo reverse transcription to generate cDNA, which shares a 10x Barcode with all cDNA from its individual cell of origin.
Resources - 3
Sequence
The resulting 10x barcoded single cell gene expression and ATAC-seq libraries are compatible with standard NGS short-read sequencing on Illumina sequencers, for massively parallel profiling of thousands of individual cells.
Resources - 4
Analyze Your Data
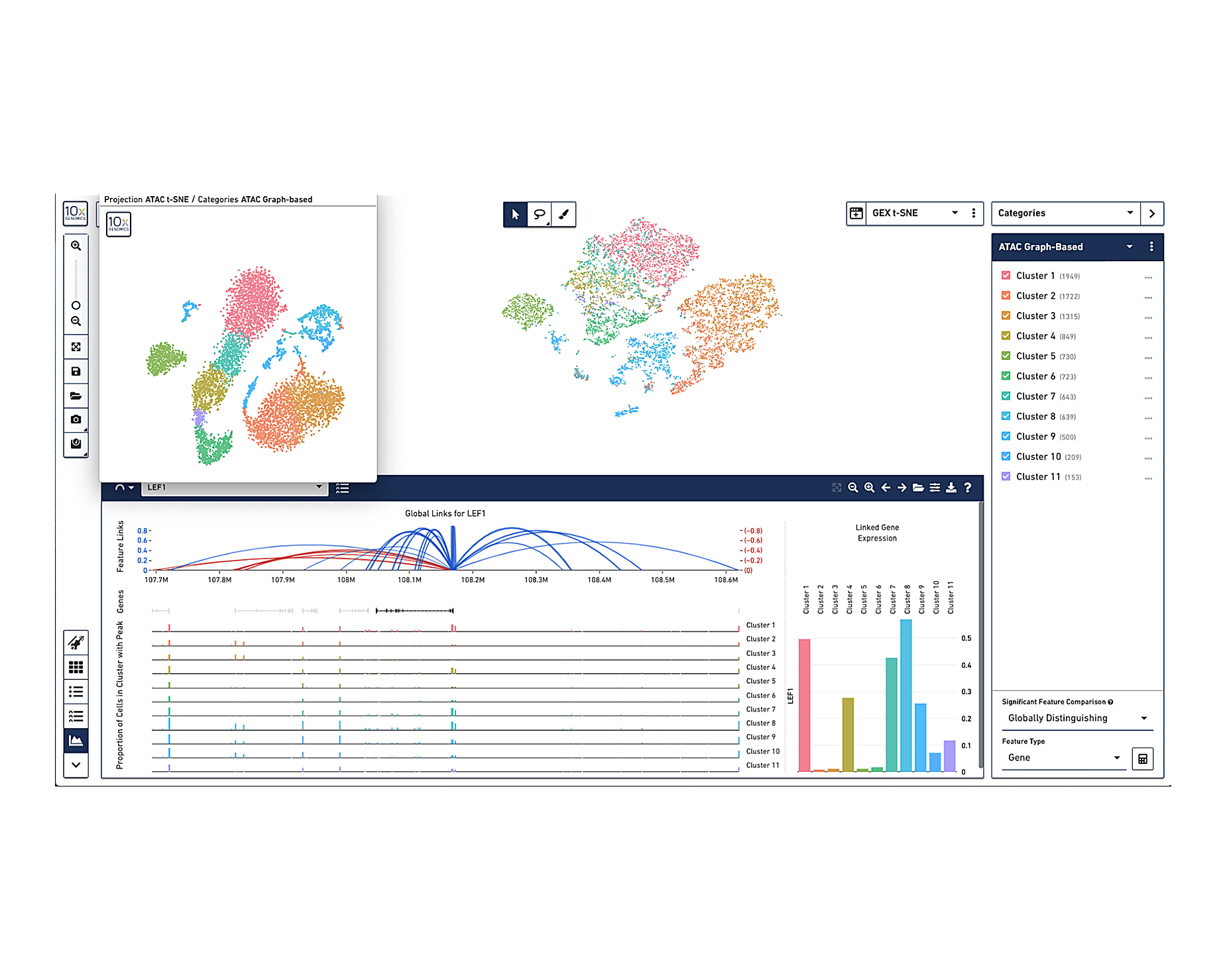
Our Cell Ranger ARC analysis software generates open chromatin and gene expression profiles for each cell, identifies clusters of cells with similar profiles, and calculates linkages between open chromatin peaks and gene expression.
Analysis pipelines output
Output includes QC information and files that can be easily used for further analysis in our Loupe Browser visualization software, or third-party R or Python tools.
Resources - 5
Visualize Your Data
Use our Loupe Browser visualization software to interactively explore your results, compare chromatin accessibility and gene expression across cell types and states, and investigate linkages between open chromatin peaks and gene expression.
Do I need to be a bioinformatician to use it?
Loupe is a point-and-click software that’s easy for anyone to download and use.
Resources
Frequently Asked Questions
It enables simultaneous profiling of the transcriptome (using 3’ gene expression) and epigenome (using ATAC-seq) from single cells to deepen your understanding of how genes are expressed and regulated across different cell types.
In the sample types tested with Single Cell Multiome ATAC + Gene Expression, performance of ATAC and gene expression is comparable to that of the corresponding standalone 10x Genomics workflow (Chromium Single Cell ATAC or Chromium Single Cell Gene Expression) when the starting input is nuclei.
No, currently Feature Barcode technology is not compatible with Single Cell Multiome ATAC + Gene Expression.
A nuclei suspension is needed. It is possible to obtain high quality nuclei suspensions from fresh and cryopreserved cells, fresh tissue, and frozen tissue. The Chromium Nuclei Isolation Kit can be used to generate high-quality nuclei suspensions from a wide range of frozen tissues with minimal optimization.
This assay has been optimized for human and mouse samples.
Yes, data from Single Cell ATAC and Single Cell Gene Expression is provided in a file format that is compatible with Single Cell Multiome ATAC + Gene Expression and can be integrated using third-party tools, such as Seurat.